6b1648806fc1de0da4b16bb23f51e0fd52065ef7
gperez2
Wed Apr 13 13:12:33 2022 -0700
making ReMap hub into native track, refs #28960
diff --git src/hg/makeDb/trackDb/human/reMap2022.html src/hg/makeDb/trackDb/human/reMap2022.html
new file mode 100644
index 0000000..8f2e448
--- /dev/null
+++ src/hg/makeDb/trackDb/human/reMap2022.html
@@ -0,0 +1,190 @@
+Description
+
+This track represents the ReMap Atlas of regulatory regions which consists of a
+large scale integrative analysis of all Public ChIP-seq data for transcriptional
+regulators from GEO, ArrayExpress and ENCODE.
+
+
+
+Below is a schematic diagram of the types of regulatory regions:
+
+- ReMap 2022 Atlas (all peaks for each analyzed datasets)
+- ReMap 2022 Non redundant peaks (merged similar target)
+- ReMap 2022 Cis Regulatory Modules
+
+
+
+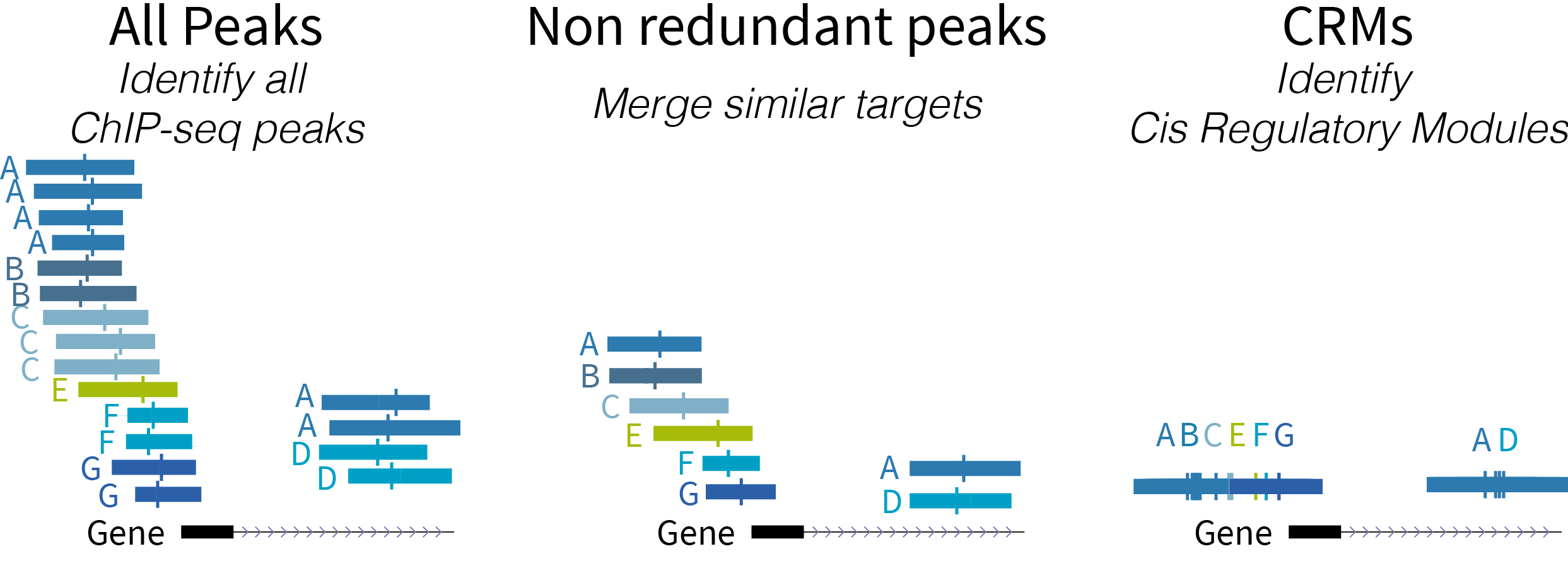 +
+
+
+ Display Conventions and Configuration
+
+-
+Each transcription factor follows a specific RGB color.
+
+-
+ChIP-seq peak summits are represented by vertical bars.
+
+-
+Hsap : A data set is defined as a ChIP/Exo-seq experiment in a given
+GEO/ArrayExpress/ENCODE series (e.g. GSE41561), for a given TF (e.g.: ESR1), in
+a particular biological condition (e.g. MCF-7).
+
Data sets are labelled with the concatenation of these three pieces of
+information (e.g. GSE41561.ESR1.MCF-7).
+
+-
+Atha : The "dataset" is defined as a ChIP-seq experiment in a given series
+(e.g. GSE94486), for a given target (e.g. ARR1), in a particular biological
+condition (i.e. ecotype, tissue type, experimental conditions ; e.g.
+Col-0_seedling_3d-6BA-4h).
+
Data sets are labelled with the concatenation of these three pieces of
+information (e.g. GSE94486.ARR1.Col-0_seedling_3d-6BA-4h).
+
+
+
+Methods
+
+This 4th release of ReMap (2022) present the analysis of a total of 8,103
+quality controlled ChIP-seq (n=7,895) and ChIP-exo (n=208) datasets from public
+sources (GEO, ArrayExpress, ENCODE). The ChIP-seq/exo datasets have been mapped
+to the GRCh38/hg38 human assembly. The "dataset" is defined as a ChIP-seq
+experiment in a given series (e.g. GSE46237), for a given TF (e.g. NR2C2), in a
+particular biological condition (i.e. cell line, tissue type, disease state or
+experimental conditions ; e.g. HELA). Datasets were labeled by concatenating
+these three pieces of information such as GSE46237.NR2C2.HELA.
+
Those merged analyses cover a total of 1,211 DNA-binding protein
+(transcriptional regulators) such as a variety of transcription factors (TFs),
+transcription co-activators (TCFs) and chromatin-remodeling factors (CRFs) for
+182 million peaks.
+
+
+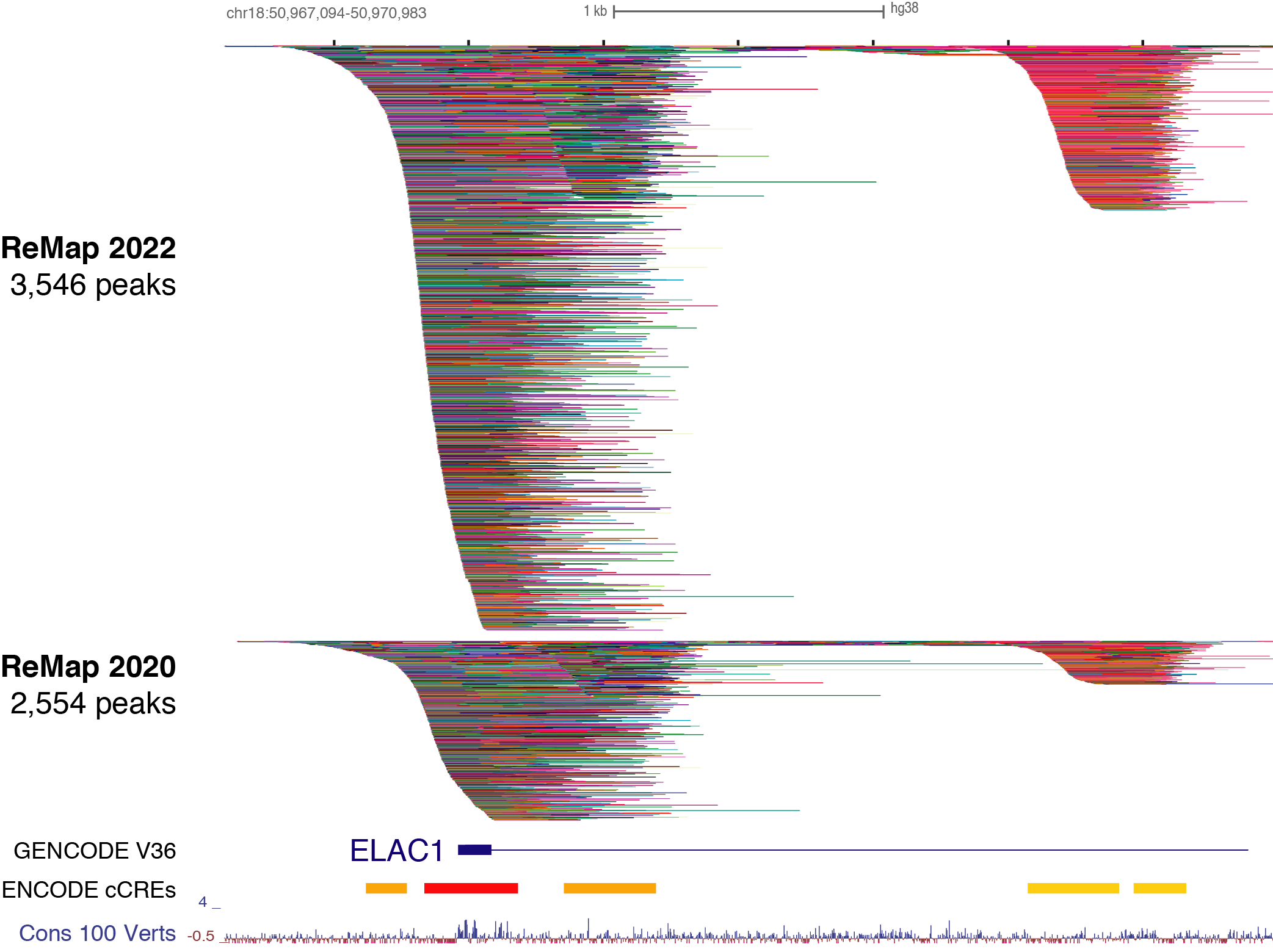 +
+
+
+GEO & ArrayExpress
+Public ChIP-seq data sets were extracted from Gene Expression Omnibus (GEO) and
+ArrayExpress (AE) databases. For GEO, the query '('chip seq' OR 'chipseq' OR
+'chip sequencing') AND 'Genome binding/occupancy profiling by high throughput
+sequencing' AND 'homo sapiens'[organism] AND NOT 'ENCODE'[project]' was used to
+return a list of all potential data sets to analyze, which were then manually
+assessed for further analyses. Data sets involving polymerases (i.e. Pol2 and
+Pol3), and some mutated or fused TFs (e.g. KAP1 N/C terminal mutation, GSE27929)
+were excluded.
+
+ENCODE Human
+Available ENCODE ChIP-seq data sets for transcriptional regulators from
+www.encodeproject.org portal were processed with the standardized ReMap pipeline.
+The list of ENCODE data was retrieved as FASTQ files from the ENCODE portal
+(https://www.encodeproject.org/) using the following filters: Assay: "ChIP-seq",
+Organism: "Homo sapiens", Target of assay: "transcription factor", Available data:
+"fastq" on 2016 June 21st. Metadata information in JSON format and FASTQ files
+were retrieved using the Python requests module.
+
+
+ChIP-seq processing
+Both Public and ENCODE data were processed similarly. Bowtie 2 (PMC3322381
+) (version 2.2.9) with options -end-to-end -sensitive was used to align all
+reads on the human genome (GRCh38/hg38 assembly). Biological and technical
+replicates for each unique combination of GSE/TF/Cell type or Biological condition
+were used for peak calling. TFBS were identified using MACS2 peak-calling tool
+(
+PMC3120977) (version 2.1.1.2) in order to follow ENCODE ChIP-seq guidelines,
+with stringent thresholds (MACS2 default thresholds, p-value: 1e-5). An input data
+set was used when available.
+
+
+Quality assessment
+To assess the quality of public data sets, a score was computed based on the
+cross-correlation and the FRiP (fraction of reads in peaks) metrics developed by
+the ENCODE Consortium (http://genome.ucsc.edu/ENCODE/qualityMetrics.html). Two
+thresholds were defined for each of the two cross-correlation ratios (NSC,
+normalized strand coefficient: 1.05 and 1.10; RSC, relative strand coefficient:
+0.8 and 1.0). Detailed descriptions of the ENCODE quality coefficients can be
+found at http://genome.ucsc.edu/ENCODE/qualityMetrics.html. The phantompeak
+tools suite was used (https://code.google.com/p/phantompeakqualtools/) to compute
+RSC and NSC.
+
+Please refer to the ReMap 2022, 2020, and 2018 publications for more details
+(citation below).
+
+
+
+
+Data Access
+
+ReMap Atlas of regulatory regions data can be explored interactively with the
+Table Browser and cross-referenced with the
+Data Integrator. For programmatic access,
+the track can be accessed using the Genome Browser's
+REST API.
+ReMap annotations can be downloaded from the
+Genome Browser's download server
+as a bigBed file. This compressed binary format can be remotely queried through
+command line utilities. Please note that some of the download files can be quite large.
+
+
+Individual BED files for specific TFs, or Cells/Biotypes or Datasets can be
+found and downloaded on the ReMap website http://remap.univ-amu.fr/ or at http://remap2022.univ-amu.fr/.
+
+
+The ReMap BED files for all version [2022, 2020, 2018, 2015] are available for
+download at the ReMap website http://remap.univ-amu.fr/ in the download tab.
+
+
+
+References
+
+
+Chèneby J, Gheorghe M, Artufel M, Mathelier A, Ballester B.
+
+ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-
+seq experiments.
+Nucleic Acids Res. 2018 Jan 4;46(D1):D267-D275.
+PMID: 29126285; PMC: PMC5753247
+
+
+Chèneby J, Ménétrier Z, Mestdagh M, Rosnet T, Douida A, Rhalloussi W, Bergon A, Lopez
+F, Ballester B.
+
+ReMap 2020: a database of regulatory regions from an integrative analysis of Human and Arabidopsis
+DNA-binding sequencing experiments.
+Nucleic Acids Res. 2020 Jan 8;48(D1):D180-D188.
+PMID: 31665499; PMC: PMC7145625
+
+
+Griffon A, Barbier Q, Dalino J, van Helden J, Spicuglia S, Ballester B.
+
+Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory
+landscape.
+Nucleic Acids Res. 2015 Feb 27;43(4):e27.
+PMID: 25477382; PMC: PMC4344487
+
+
+Hammal F, de Langen P, Bergon A, Lopez F, Ballester B.
+
+ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an
+integrative analysis of DNA-binding sequencing experiments.
+Nucleic Acids Res. 2022 Jan 7;50(D1):D316-D325.
+PMID: 34751401; PMC: PMC8728178
+
+
 +
+ +
+